Our paper showing that the sexual selection can both inflate or deflate MHC gene number is in the wild!
26 Feb 2020Some time ago, I did a post-doc in the Evolutionary Biology Group, a lab run by prof Jacek Radwan at Adam Mickiewicz University in Poznań. I was working on the evolution of copy number variation of the major histocompatibility complex (MHC) genes. MHC genes code for molecules binding fragments of proteins from both self and pathogens thus constituting one of the elements of the immune system. They are found in all jawed vertebrates. MHC proteins can also ‘recognize’ other individuals of the same species (hence problems with organ transplants), and they are known to impact mating preference in animals (including humans). Our work focused on the question of how different mating rules:
- Preference for mates with the smallest number of identical MHC variants
- Preference for mates with a minimal proportion of shared identical MHC variants within the sum of variants in both partners
- Preference for mates with the largest number of MHC variants different from the chooser
- No preference, just random matching of mates
will impact an individual’s number of MHC variants? It turns out our simulations showed that mating preferences for MHC dissimilarity might have significant consequences for the evolution of MHC gene family expansion. Differences in specific mechanisms of mating rules can lead to very different outcomes and have the potential to explain interspecific differences in the number of MHC genes.
Abstract
Major histocompatibility complex (MHC)-based mating rules can evolve as a way to avoid inbreeding or to increase offspring immune competence. While the role of mating preference in shaping the MHC diversity in vertebrates has been acknowledged, its impact on individual MHC diversity has not been considered. Here, we use computer simulations to investigate how simple mating rules favouring MHC-dissimilar partners affect the evolution of the number of MHC variants in individual genomes, accompanying selection for resistance to parasites. We showed that the effect of such preferences could sometimes be dramatic. If preferences are aimed at avoiding identical alleles, the equilibrium number of MHC alleles is much smaller than under random mating. However, if the mating rule minimizes the ratio of shared to different alleles in partners, MHC number is higher than under random mating. Additionally, our simulations revealed that a negative correlation between the numbers of MHC variants in mated individuals can arise from simple rules of MHC-disassortative mating. Our results reveal unexpected potential of MHC-based mating preferences to drive MHC gene family expansions or contractions and highlight the need to study the mechanistic basis of such preferences, which is currently poorly understood.
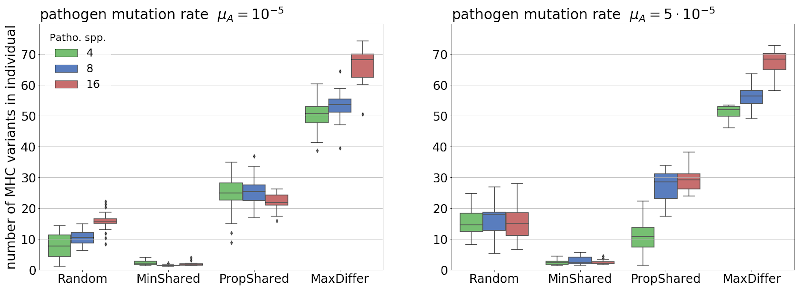 Fig.1. The effect of MHC-based mating rules minimising the number (MinShared), minimising the proportion (PropShared) of shared variants, and maximising the number of different variants (MaxDiffer), on the number of unique MHC variants in an individual, compared to random mating (Random).
Fig.1. The effect of MHC-based mating rules minimising the number (MinShared), minimising the proportion (PropShared) of shared variants, and maximising the number of different variants (MaxDiffer), on the number of unique MHC variants in an individual, compared to random mating (Random).
The paper was published in the Proceeding of the Royal Society B: Biological Sciences.
Cite:
The code of this project can be found at my GitHub repository.
